Introduction
There is a saying that our life is determined by our genes, and along with the adage that “we are born with it”. Our genes may dictate the blueprint for physical traits, predispositions to certain diseases, and even traits like intelligence, but our lifestyle and environment can also influence our development and the choices we make. While we cannot change our genes, we can shape our lives by choosing the right environment and lifestyle.
Or is it?
A 2012 study conducted by psychologists at the University of Edinburgh suggests that genes play an important role in a person’s predisposition toward determination, sociability and self-control, and ultimately, success in life. The researchers explained that genetically determined traits such as self-control, decisiveness or sociability can make the difference between success and failure. Does this mean that winners are born and success in life is determined at birth?
What about the proverb – nature versus nurture? Scientists have long debated the importance of nature and nurture – genes and environment – in the choices people make and the paths they take in life. However, it is now widely accepted that both nature and environment play important roles in the development of a person’s personality, behaviour, and abilities. Recent research [1] has found that the genetics of a person’s friends and schoolmates influence how long they stay in school, which makes perfect sense – after all, people seek out the company of friends with similar interests and hobbies.
The study suggests that our genes have a direct impact on the environment we live in; we are the company we keep. For example, if your friend is diligent and completes his or her assignments on time, that could make the group more academically compliant and explain how one group of students may be treated better than others because of observable behaviours that have individual genetic origins but play a social role because they elicit a positive response from the teacher. And this better treatment may contribute to better performance that affects all members of the group independently. The opposite is also true: A 2018 study [2] showed that smoking, drugs, and other risky behaviours can cause negative developmental outcomes. People with certain genes associated with higher levels of education are always less likely to have genes associated with smoking and to be smokers. The social environment may influence the extent to which certain genes affect the association between behaviours.
Now which affects which?
These traits and characteristics play an important role in how life progresses, if you are successful in career and have healthy relationships, or not. If that’s so, then what do the genes do and where does the traits come from? [3]. DNA affects how well exercise affects us (some of us are truly fated to be fat..) [4], how tall we will be (some of us a truly fated to be short) [5], our happiness [6] and even how long an organisms will live, some species of jellyfish are immortal [7]! How attractive an individual turns out to be or why some others are plagued with numerous genetic diseases. For example the OCA2 gene codes the appearance of blue eyes or rather the lack of the protein melanin. That’s it, some people are indeed just born beautiful.
However, there’s a problem with just looking at genes.
The amoeba Proteus is a tiny microbe that is barely visible to the human eye. It can grow up to 1 mm long, but its average size is 250-750 micrometers. It has only 290 billion base pairs, making it 100 times larger than the human genome. The 3 GB human genome is thought to contain fewer than 30,000 genes, perhaps as few as 23,000, which are necessary for programming the human body [8]. Why does such a small organism need so many genes and a large complex organism like humans so few in comparison?
Did you know that as humans we share about 60% of our genes with a banana and a chicken? Humans also have a 71.4% genetic similarity to the zebrafish [9]. What makes us look the way we look and not like a chicken?
Clearly the number of base pairs in DNA is not an accurate indicator.
This brings us to the scientific discipline of genomics. We now know that genes play a greater role in traits as self-control, decision making, or sociability than previously thought. DNA has been used to disprove homosexuality or sexual preferences is not genetic [10].
Genetic determinism is the idea that genes have a controlling influence on human health, behaviour, and disease. However, the current view is that genes have a complex relationship with environmental influences, which means that genes alone are not determinative. The field of genomics is vast. There are sub-experts of experts of experts in sub-fields of genomics, see Figure 2 to see what comes after genomics.
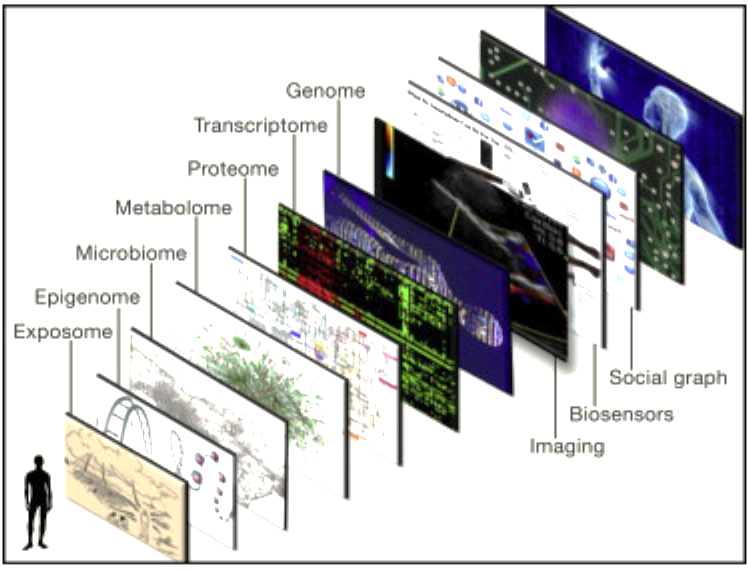
The fact that each of us is truly biologically unique, which is true even for identical twins, is not fully appreciated. Now that it is possible to perform a comprehensive “omic” assessment of an individual, including DNA and RNA sequence and at least some characterization of the proteome, metabolome, microbiome, autoantibodies, and epigenome, it has become abundantly clear that each of us truly has a unique biological content. Far beyond the appeal of the incomparable fingerprint or snowflake concept, this unique, individual data and information offers a remarkable and unprecedented opportunity to improve medical treatment and develop preventive strategies to maintain health.
There are numerous other factors affecting one’s genes – I’ve decided to talk briefly about polymorphism, pleiotropy, polygenic traits and epigenetics just to name a few. Let us break down what some of these big words mean.
Polymorphism, in biology, is discontinuous genetic variation that results in different individuals among members of a single species. Pleiotropy describes genes that have multiple effects on an organism. For example, a single gene can affect both eye color and susceptibility to disease. The term “polygenic” refers to the inheritance of traits or characteristics that are determined by more than one gene. This means that multiple genes contribute to the expression of a particular trait, rather than just one gene. Polygenic inheritance can be observed in many human traits, such as height, eye color, and skin color.
If this is not confusing enough, we should briefly mention epigenetics, which is the influence of environmental or external factors that lead to phenotypic changes without altering the DNA sequence itself.
Earlier we described co-influences of sociological influences by one’s genes having a causal factor to academic success [1], now research shows that epigenetic factors could also be involved [12], so now it could be the original genes – i.e. the “nature” is thus shaped by nuturing [13].
Remember how earlier I lamented that some of us are truly born to be fat [4], short [5] and sad [6]? There is good news! Science has found the source of cravings – Ghrelin, a small peptide released from the stomach, is an orexigenic hormone that regulates food intake, body weight and glucose homeostasis. Behavioral studies show that ghrelin is implicated in the regulation of both hedonic and homeostatic feeding [14].
We now know what makes us sad and what makes us eat too much in the hope of suppressing the sadness, but this makes us fat, which is then embarrassing in society, and we become even sadder. It’s a vicious genetic cycle. Blame it on our genes.
That each of us is truly biologically unique, extending to even monozygotic, “identical” twins, is not fully appreciated. Now that it is possible to perform a comprehensive “omic” assessment of an individual, including one’s DNA and RNA sequence and at least some characterization of one’s proteome, metabolome, microbiome, autoantibodies, and epigenome, it has become abundantly clear that each of us has truly one-of-a-kind biological content because of external factors.
Further understanding how to change our own genetic code gives us humans a remarkable and unprecedented opportunity to improve medical treatment and develop preventive strategies to preserve health. This new exciting frontier is referred to as Individualized genetic medicine [11] and the technology to do gene editing now exists – CRISPR [15-18].
I’ve always wondered about the genetic effects and how we can use this knowledge to improve disease treatment and therapeutics, so I thought it would be fun to give R Programming a shot.
R is a statistical programming language and R-studio works in a similar manner to MATLAB – except it’s open-source and comes with a ton of tools and libraries (similar to MATLAB toolboxes). I got R-studio running and after a few YouTube tutorials such as this and that. I have a study plan in mind done.
The study
The study is simple. This is an amateur bioinformatics study to identify genomic signatures with polymorphic and pleiotropic associations for hypertension and diabetes. Cardiovascular and metabolic diseases, also referred to as “cardiometabolics” represent an increasing disease burden of epidemic proportions. Hypertension affects 30% of the world’s population whilst Type-2 diabetes mellitus (DM -2), affects up to 10% of the world’s adult population.
Many genomic-proteomic-metabolomic traits have been identified as contributing or predictive factors for the development of cardiometabolic diseases. Prediction of disease based on genotyping alone has resulted in poor associations because of unexpected multinomic interactions. Pleiotropic, cross-boundary biomolecular interactions between gene regulation and protein expression, as well as transport of metabolites that affect biological functions, are difficult to explore to achieve a better understanding of disease outcomes [19].
Due to the enormous size and scope of the human genome database, it is a technical challenge to aggregate all multinomic datasets and different data formats and to represent the different multinomic levels. Therefore, genome-wide association studies (GWAS) are a powerful method by which multinomic cardiometabolic relationships for pleiotropic and polymorphic behaviour could be identified [20-24]. The complex cardiometabolic genomic-proteomic interactions are of significant medical and academic interest in the search for novel therapeutics [25-27].
Method
The aim of this study is to apply gene set enrichment analysis (GSEA) techniques in R-Studio to different datasets to identify potential polymorphic and pleiotropic relationships. The analyses and illustrations were generated using various R library packages in R-Studio. I looked through several genetic databases such as Genecards, GenBank, Kyoto Encyclopaedia of Genes and Genomes (KEGG) and eventually settled on Gene Expression Omnibus (GEO) and GWAS catalog.
Gene Set Enrichment Analysis (GSEA) was performed on the hypertension and diabetes dataset taken directly from the GEO database and GWAS catalogue, using search terms for four disease states: “cardiovascular disease,” “type diabetes II “, “hypertension,” and “obesity”.
The approach emulates the GWAS relationship between four diseases – dilipidaemia, obesity, DM-2 and hypertension conducted by the authors in [28]. The dataset was further queried for trait IDs of the sample and the list of significant association were downloaded and formatted as a supplementary material to this report as “GSEADM2.csv”, “GSEAhypertension.csv”, “GSEAcardiovascular disease.csv” and “GSEAobesity.csv”. Table 1 characterizes the dataset and resource from the GWAS catalog for this GSEA study.
Table 1 Characterization of the selected datasets from GWAS catalog
| Traits | Trait ID | Studies | Associations | P-value cutoff | No. of genes | No. of SNPs |
| Cardiovascular disease (CVD) | EFO_0000319 | 615 | 6454 | 9 x 10-6 | 3530 | 5386 |
| Diabetes Mellitus (Type 2) | EFO_0001360 | 173 | 4049 | 9 x 10-6 | 1810 | 3149 |
| Hypertension | EFO_0000537 | 89 | 731 | 9 x 10-6 | 452 | 674 |
| Obesity | EFO_0001073 | 36 | 361 | 9 x 10-6 | 258 | 326 |
Single nucleotide polymorphisms (SNPs) (pronounced “snips”) are the most common type of genetic variation in humans. Each SNP represents a difference in a single DNA building block called a nucleotide. For example, a SNP can replace the nucleotide cytosine (C) with the nucleotide thymine (T) in a particular section of DNA.
SNPs normally occur everywhere in a person’s DNA, once for every 1,000 nucleotides, which means there are about 4 to 5 million SNPs in a person’s genome. These variations occur in many people; to be classified as an SNP, a variant must be found in at least 1 percent of the population. To date, researchers have found more than 600 million SNPs in populations worldwide.
SNPs are commonly found in DNA between genes, acting as biological markers associated with a particular disease. When SNPs occur within a gene or in a regulatory region near a gene, they directly affect the genetic function or variant of a disease.
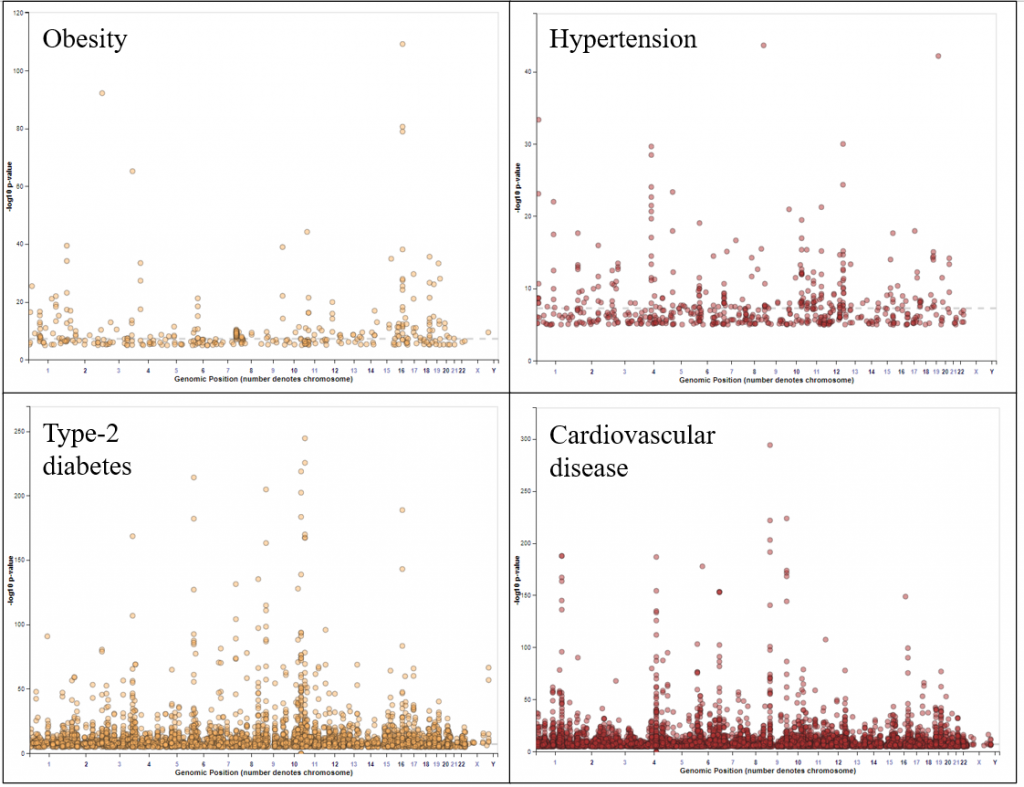
I then filtered the variant IDs for gene names or gene symbols and then analysed for gene set enrichment analysis in Enrichr [29]. The Manhattan plots of the SNP association of the four diseases have been shown Figure 4 and due to the size of the dataset, the analysis doesn’t reveal significant observations.
Results
GWAS analysis identified 312 genes for obesity, 523 genes for hypertension, 2614 genes for DM -2, and 4 917 genes for cardiovascular disease (CVD). There were some overlapping gene codes that were removed. But the results are dramatic.
Omg, 2,614 genes can cause diabetes and 4917 genes can cause cardiovascular disease. It looks like humanity is predisposed to get these two diseases. Life is unfair.
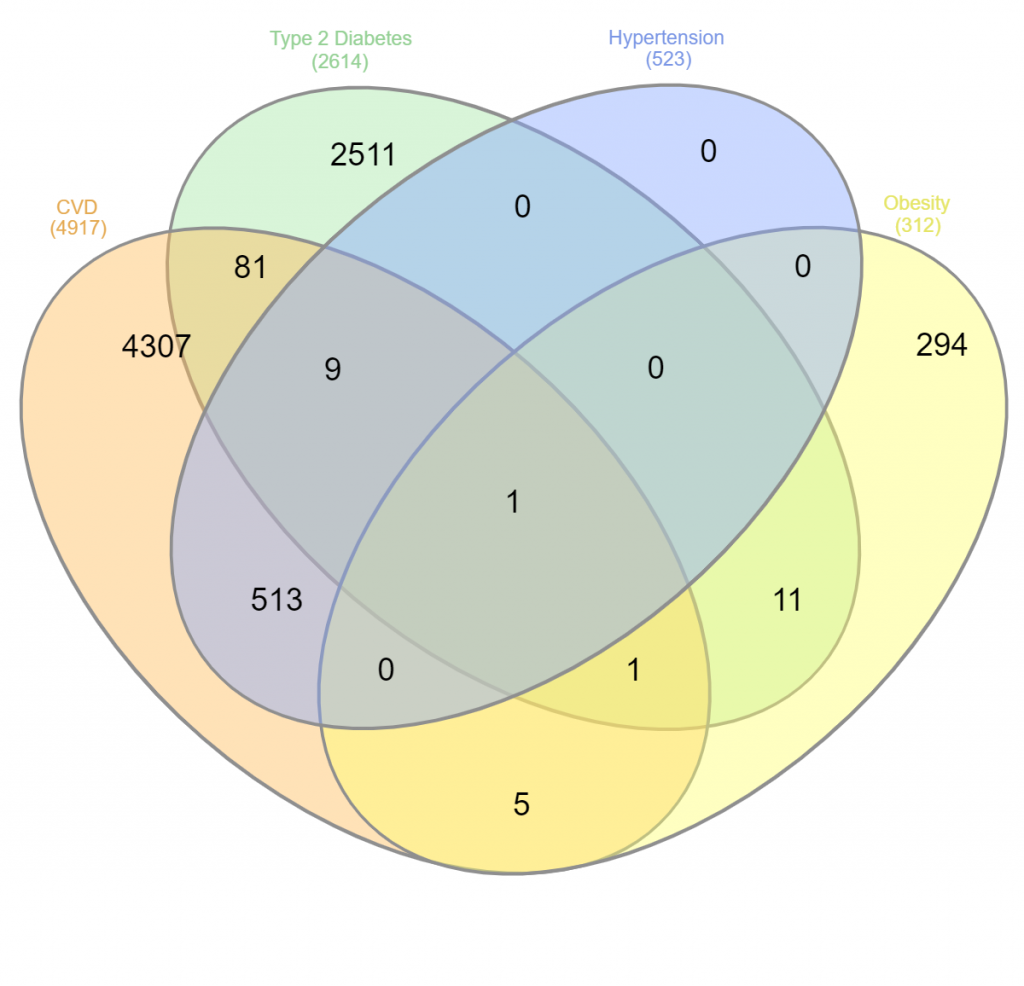
While numerous genes are found to affect two disease groups, the number of pleiotropically associated genes decreases significantly-10 genes affect 3 diseases (CVD, DM -2, hypertension) and only 1 single gene affects 4 diseases (CVD, DM -2, obesity, and hypertension). Based on the data set entered, the data are presented in a Venn diagram in which the clearly overlapping areas represent the relationship between hypertension, diabetes, cardiovascular disease, and obesity. Figure 5 is a Venn diagram showing the relationship between the data sets, with each colour representing a condition factor.
Genetic predisposition to the development of diabetes and hypertension is well established [30, 31], but conclusive evidence remains elusive. For example, of the 43 monogenic variants associated with hypertension, none has been linked to diabetes – a metabolic disease [32].
However, in another example, obesity was associated with RPGRIP1L genes [33], Melanocortin 4 receptor (MC4R) deficiency [34], and GYS2 glycogen synthase [25] with further network analysis identifying common obesity-related genes [35].
The final 9 genes causing three diseases are shown as purple text in Table 2 and as a purple circle in Figure 5. The results of the study are not surprising and appear to be consistent with those reported by [28]. For example, APOE (apolipoprotein E) is known to contribute to lipid metabolism in the mammalian body. One subtype is involved in Alzheimer’s disease and cardiovascular disease. TRIB3 is associated with at least two diseases (DM and hypertension) [36] and ADCY3 can polymorph diabetes, hypertension and obesity [28, 37].
The single gene ” SUGP1-rs10401969″ has been identified as causing CVD, DM -2 and obesity and is shown as orange text in Table 2 and as an orange circle in Figure 5. SUGP1-rs10401969 has been identified as a regulator of cholesterol metabolism and thus may be associated with the phenotype of CVD, type 2 diabetes, and obesity; it has recently been studied as a regulator of cholesterol metabolism [38].
The most interesting finding was that the single gene FTO-rs1421085, which affects all four diseases, was recently associated with increased body weight [39], this gene is shown in Table 2 as red text and in Figure 5 as a red circle.
Table 2 Final list of polymorphic / pleiotropic genes that have been found to cause a combination of CVD, DM-2, hypertension and/or obesity
| Gene | Full name | SNP | CVD | DM-2 | Hypertension | Obesity |
| TRIB3 | Tribbles homolog 3 | rs2295490 | – | ✓ | ✓ | – |
| ADCY3 | Adenylate Cyclase 3 | rs750852737 | – | ✓ | ✓ | ✓ |
| APOE | Apolipoprotein E | rs429358 | ✓ | ✓ | ✓ | – |
| HECTD4 | HECT domain E3 ubiquitin protein ligase 4 | rs2074356& rs77768175 | ✓ | ✓ | ✓ | – |
| LINC01405 | Long intergenic non-protein coding RNA 1405 | CUX2-rs12229654 | ✓ | ✓ | ✓ | – |
| NME9 | Thioredoxin domain-containing protein 6 | MRAS-rs4678408 | ✓ | ✓ | ✓ | – |
| OAS1 | 2′-5′-Oligoadenylate Synthetase 1 | AC004551.1-rs11066453 | ✓ | ✓ | ✓ | – |
| PURG | Purine Rich Element Binding Protein G | rs2725371 | ✓ | ✓ | ✓ | – |
| SLC39A8 | Solute Carrier Family 39 Member 8 | rs13107325 | ✓ | ✓ | ✓ | – |
| UMOD | Uromodulin and minor G allele | rs13333226 | ✓ | ✓ | ✓ | – |
| SUGP1 | SURP And G-Patch Domain Containing 1 | rs10401969 | ✓ | ✓ | – | ✓ |
| FTO | Alpha-Ketoglutarate Dependent Dioxygenase | rs1421085 | ✓ | ✓ | ✓ | ✓ |
I didn’t expect this, but this study concludes that hypertension is statistically associated with heart attacks, but the genomic data don’t show a clear association with diabetes. As for the GSEA analysis, although the study finds 9 different genes that could cause both hypertension and diabetes, only one gene “FTO-rs1421085”, the fat mass and obesity-associated protein (FTO) gene, overlaps with all four disease states. This finding suggests further investigation as a potential candidate for gene therapy therapeutics.
Conclusion
GWAS research has uncovered overlapping cardiometabolic relationships for cardiovascular disease and DM [21] and is the most promising approach to uncovering pleiotropic relationships between biomarkers and risk factors for hypertension and diabetes [28, 40].
In my amateur bioinformatics study, I found an overlapping gene, FTO, an enzyme that has been uniquely linked to obesity in humans [41-43] by suppressing mitochondrial thermogenesis in adipocyte progenitor cells. This finding suggests further investigation of the FTO gene as a potential candidate for CRISPR-like therapeutics to treat diabetes and hypertension
However, the etiology of both diseases (DM and hypertension) in cardiometabolic diseases is extremely complex due to pleiotropic and polymorphic relationships and requires further investigation
Imagine a world where we can selectively turn genes on and off to create a stronger or better human? Humans with no diseases? Humans with animal-like characteristics?
Hmmm?
Disclaimer
This study, the experiments, the collection of genotype and phenotype information, the statistical and bioinformatic analysis, and the manuscript were performed solely by me. Please note that I am not an expert in bioinformatics and that all analyzes and methods described in this report have not been peer reviewed or examined for scientific accuracy.
References
1. Domingue, B.W., et al., The social genome of friends and schoolmates in the National Longitudinal Study of Adolescent to Adult Health. Proceedings of the National Academy of Sciences, 2018. 115(4): p. 702-707.
2. Wedow, R., et al., Education, smoking, and cohort change: Forwarding a multidimensional theory of the environmental moderation of genetic effects. American Sociological Review, 2018. 83(4): p. 802-832.
3. Buchanan, A.V., et al., What are genes “for” or where are traits “from”? What is the question? Bioessays, 2009. 31(2): p. 198-208.
4. Chung, H.C., et al., Do exercise-associated genes explain phenotypic variance in the three components of fitness? a systematic review & meta-analysis. PLOS ONE, 2021. 16(10): p. e0249501.
5. Wood, A.R., et al., Defining the role of common variation in the genomic and biological architecture of adult human height. Nature genetics, 2014. 46(11): p. 1173-1186.
6. Archontaki, D., G.J. Lewis, and T.C. Bates, Genetic Influences on Psychological Well-Being: A Nationally Representative Twin Study. Journal of Personality, 2013. 81(2): p. 221-230.
7. Pascual-Torner, M., et al., Comparative genomics of mortal and immortal cnidarians unveils novel keys behind rejuvenation. Proceedings of the National Academy of Sciences, 2022. 119(36): p. e2118763119.
8. Hornyak, G.L., et al., Introduction to nanoscience and nanotechnology. 2008, CRC press. p. 771.
9. Howe, K., et al., The zebrafish reference genome sequence and its relationship to the human genome. Nature, 2013. 496(7446): p. 498-503.
10. Ganna, A., et al., Large-scale GWAS reveals insights into the genetic architecture of same-sex sexual behavior. Science, 2019. 365(6456): p. eaat7693.
11. Topol, Eric J., Individualized Medicine from Prewomb to Tomb. Cell, 2014. 157(1): p. 241-253.
12. Seebacher, F. and J. Krause, Epigenetics of Social Behaviour. Trends in Ecology & Evolution, 2019. 34(9): p. 818-830.
13. Powledge, T.M., Behavioral Epigenetics: How Nurture Shapes Nature. BioScience, 2011. 61(8): p. 588-592.
14. Serrenho, D., S.D. Santos, and A.L. Carvalho, The Role of Ghrelin in Regulating Synaptic Function and Plasticity of Feeding-Associated Circuits. Frontiers in Cellular Neuroscience, 2019. 13.
15. Barrangou, R. and J.A. Doudna, Applications of CRISPR technologies in research and beyond. Nature biotechnology, 2016. 34(9): p. 933-941.
16. Kaminski, M.M., et al., CRISPR-based diagnostics. Nature Biomedical Engineering, 2021. 5(7): p. 643-656.
17. Lino, C.A., et al., Delivering CRISPR: a review of the challenges and approaches. Drug delivery, 2018. 25(1): p. 1234-1257.
18. Terns, M.P. and R.M. Terns, CRISPR-based adaptive immune systems. Current opinion in microbiology, 2011. 14(3): p. 321-327.
19. Baum, K., J.C. Rajapakse, and F. Azuaje, Analysis of correlation-based biomolecular networks from different omics data by fitting stochastic block models. F1000Research, 2019. 8.
20. Solovieff, N., et al., Pleiotropy in complex traits: challenges and strategies. Nature Reviews Genetics, 2013. 14(7): p. 483-495.
21. Atanasovska, B., et al., GWAS as a driver of gene discovery in cardiometabolic diseases. Trends in Endocrinology & Metabolism, 2015. 26(12): p. 722-732.
22. Taraszka, K., N. Zaitlen, and E. Eskin, Leveraging pleiotropy in genome-wide association studies in multiple traits with per trait interpretations. bioRxiv, 2020: p. 2020.05.17.100172.
23. Yang, J.J., L.K. Williams, and A. Buu, Identifying pleiotropic genes in genome-wide association studies from related subjects using the linear mixed model and Fisher combination function. BMC Bioinformatics, 2017. 18(1): p. 376.
24. Xiang, R., et al., Effect direction meta-analysis of GWAS identifies extreme, prevalent and shared pleiotropy in a large mammal. Communications Biology, 2020. 3(1): p. 88.
25. Sookoian, S. and C.J. Pirola, Genetics of the cardiometabolic syndrome: new insights and therapeutic implications. Therapeutic advances in cardiovascular disease, 2007. 1(1): p. 37-47.
26. Rodriguez-Oquendo, A., Translational cardiometabolic genomic medicine. 2015: Academic Press.
27. Sattar, N., M.V. Holmes, and D. Preiss, Research digest: genetics in cardiometabolic disease. The Lancet Diabetes & Endocrinology, 2018. 6(12): p. 922.
28. Sanghera, D.K., et al., Obesity genetics and cardiometabolic health: Potential for risk prediction. Diabetes, Obesity and Metabolism, 2019. 21(5): p. 1088-1100.
29. Chen, E.Y., et al., Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC bioinformatics https://maayanlab.cloud/Enrichr/, 2013. 14(1): p. 1-14.
30. Qi, Q., et al., Genetic predisposition to high blood pressure associates with cardiovascular complications among patients with type 2 diabetes: two independent studies. Diabetes, 2012. 61(11): p. 3026-3032.
31. Sowers, J.R., Diabetes mellitus and vascular disease. Hypertension, 2013. 61(5): p. 943-947.
32. Ehret, G.B. and M.J. Caulfield, Genes for blood pressure: an opportunity to understand hypertension. European heart journal, 2013. 34(13): p. 951-961.
33. Speakman, J.R., The ‘fat mass and obesity related’(FTO) gene: mechanisms of impact on obesity and energy balance. Current obesity reports, 2015. 4(1): p. 73-91.
34. Farooqi, I.S., et al., Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. New England Journal of Medicine, 2003. 348(12): p. 1085-1095.
35. Su, L.-n., et al., Network analysis identifies common genes associated with obesity in six obesity-related diseases. Journal of Zhejiang University-SCIENCE B, 2017. 18(8): p. 727-732.
36. Zhou, J., et al., Polytropic influence of TRIB3 rs2295490 genetic polymorphism on response to antihypertensive agents in patients with essential hypertension. Frontiers in pharmacology, 2019. 10: p. 236.
37. Saeed, S., et al., Loss-of-function mutations in ADCY3 cause monogenic severe obesity. Nature genetics, 2018. 50(2): p. 175-179.
38. Kim, M.J., et al., SUGP1 is a novel regulator of cholesterol metabolism. Human molecular genetics, 2016. 25(14): p. 3106-3116.
39. Hebbar, P., et al., FTO Variant rs1421085 Associates With Increased Body Weight, Soft Lean Mass, and Total Body Water Through Interaction With Ghrelin and Apolipoproteins in Arab Population. Frontiers in Genetics, 2020. 10.
40. Whitfield, J.B., Genetic insights into cardiometabolic risk factors. The Clinical Biochemist Reviews, 2014. 35(1): p. 15.
41. Loos, R.J. and G.S. Yeo, The bigger picture of FTO—the first GWAS-identified obesity gene. Nature Reviews Endocrinology, 2014. 10(1): p. 51-61.
42. Claussnitzer, M., et al., FTO obesity variant circuitry and adipocyte browning in humans. New England Journal of Medicine, 2015. 373(10): p. 895-907.
43. Katus, U., et al., Association of FTO rs1421085 with obesity, diet, physical activity, and socioeconomic status: A longitudinal birth cohort study. Nutr Metab Cardiovasc Dis, 2020. 30(6): p. 948-959.






